What is NABEECO? (for reviewers only)
NABEECO, is a software tool for solving the network alignment problem. NABEECO stands for Network Alignment with BEE Colony Optimization and it utilizes the artificial bee colony computing strategies for solving the so-called Graph Edit Distance problem.What is Network Alignment?
| Network Alignment is a computationally hard bioinformatics problem. It aims to find the "best" one-to-one correspondence between nodes from two input networks without using any external knowledge, such as sequence similarity. In NABEECO the "best" alignment is the one, which has the lowest graph editing cost. Applied to Protein-Protein Interaction (PPI) networks, Network Alignment, finds its applications in the validation of interactions and the prediction of new ones, as well as for extracting conserved subnetworks in a set of given PPI networks. Network Alignment is a novel basis, in addition to DNA and amino acid sequence similarity, for further interspecies network analysis in drug development, including (functional) knowledge transfer between species. |
Illustration of GED
|
Transforming graph G1 graph into G2 delete edge e1 delete node n1 delete edge e2 insert edge e4 insert node n2 insert edge e3. Cost of edit operations: deletion/insertion of edges/nodes – 1, substitution of nodes/edges – 0. GED(G1, G2)=6 |
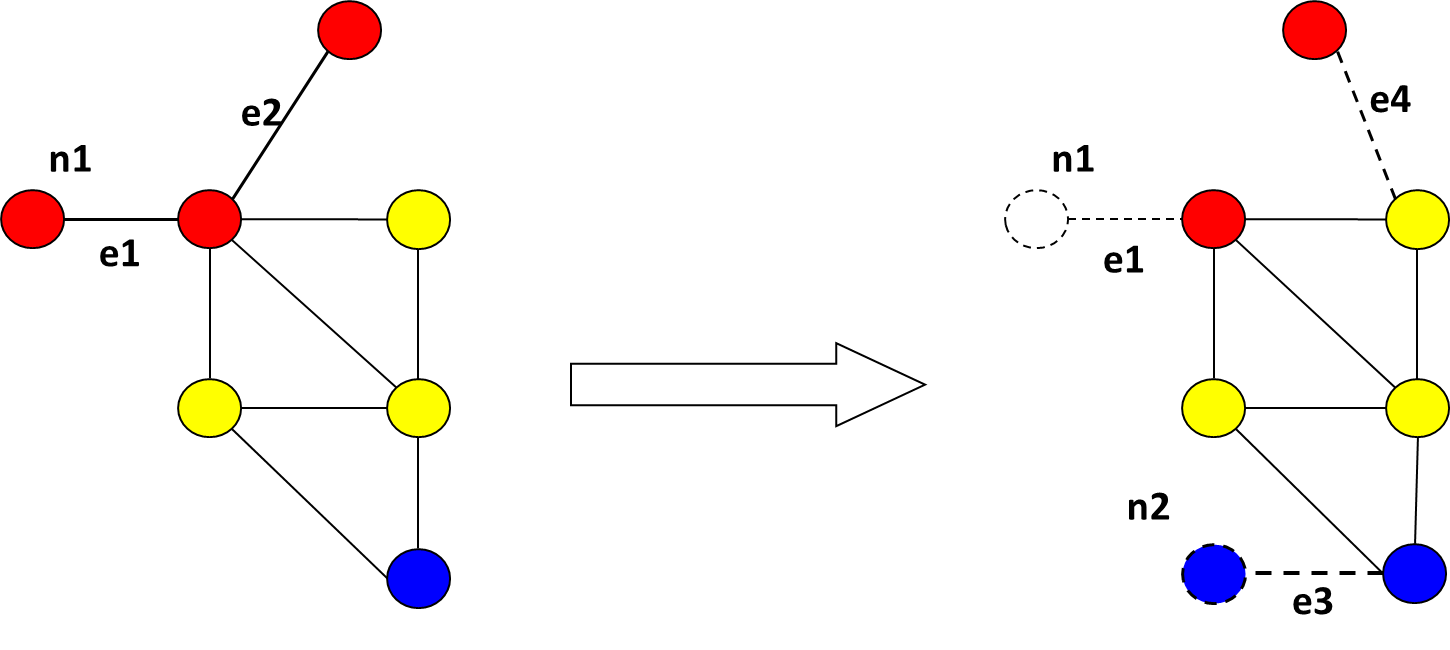
|
Illustration of Artificial Bee Colony
| Artificial bee colony consists of two types of bees: scouts and workers. Scouts are to explore randomly new sources of food. Workers explore local neighborhood of known food sources. The information about the food source and amount of food it posses is shared in the hive. The more food contains the source the higher its chance to be picked up as target to fly to. |
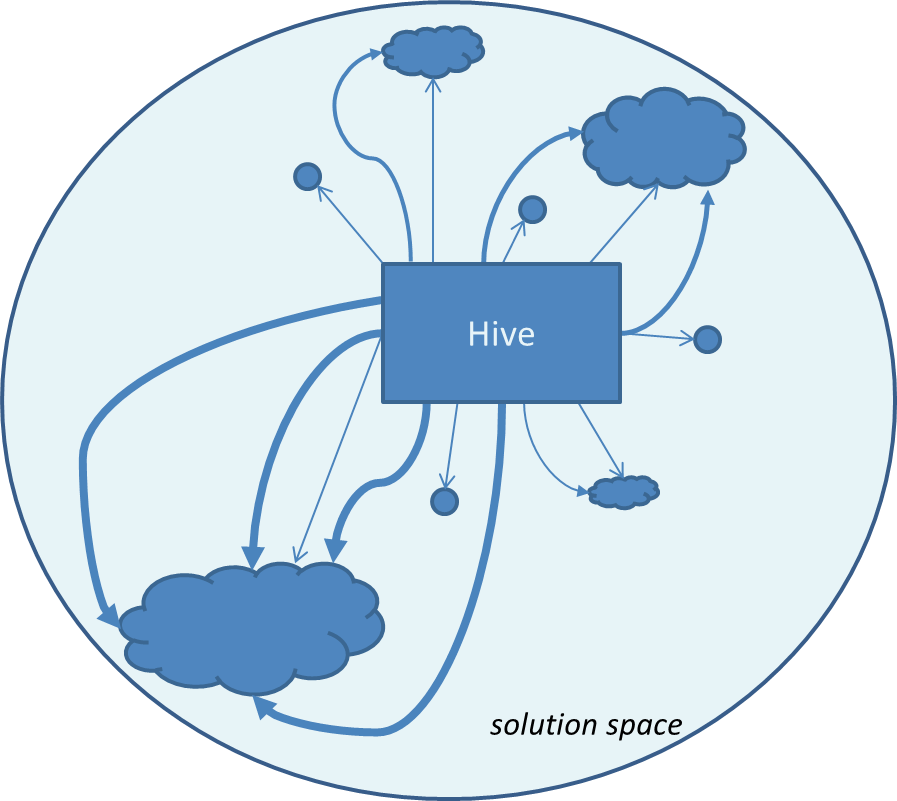
|
Download
Here are the links to download NABEECO:README.txt
linux64
win32
freebsd64
data
example.sh
How to run NABEECO?
- Start ./nabeeco (or nabeeco_win32.exe for Windows) with no parameters for a short description of the command line parameters.
- See also example.sh for an example of NABEECO with set parameters.
- NABEECO accepts networks in .ntw and .sif formats (See README file for details).
- See also example.sh for an example of NABEECO with set parameters.
- Command line example:
./nabeeco --save example_out --no-save --groups mCMV EBV --ntw data/mCMV.ntw --ntw data/EBV.ntw --grsig data/mCMV.sigs mCMV --grsig data/EBV.sigs EBV --pop 200 --maxsame 300 --no-prematch --no-workfiles --undirected
Citation
[GECCO 2013] Rashid Ibragimov, Jan Martens, Jiong Guo, Jan Baumbach. NABEECO: Biological Network Alignment with Bee Colony Optimization Algorithm; In: Proc. of Genetic and Evolutionary Computation Conference (GECCO 2013), to appear.Where was NABEECO developed?
NABEECO was mainly developed at the Max-Planck-Institut für Informatik in SaarbrückenVisit also LINK1 and LINK2 for further publications of our group

